The probiotic industry has long-faced challenges in accurately enumerating viable cells in their products. The traditional “gold standard” method, plate count, often falls short in providing precise data due to the high demands of quantification and identification. This is primarily because cultivation relies on the assumption that each colony arises from a single bacterial cell, which may not always be the case. Additionally, viable but non-culturable (VBNC) cells might not form colonies on agar plates but can still multiply and exert their probiotic effects in the human colon.
It is crucial to understand that cultivation represents an artificial environment for bacteria. While it was a groundbreaking method over 100 years ago, today’s advanced techniques in modern microbiology offer much more accurate and reliable means of analysis than simple cultivation.
A new and powerful method, called Flow-FISH, was published in June 2024 and represents a significant leap forward in the quality control of probiotic products. This innovative technique offers significant advantages over conventional methods by identifying and quantifying only living bacterial cells in a probiotic product.
The Pillars of Flow-FISH
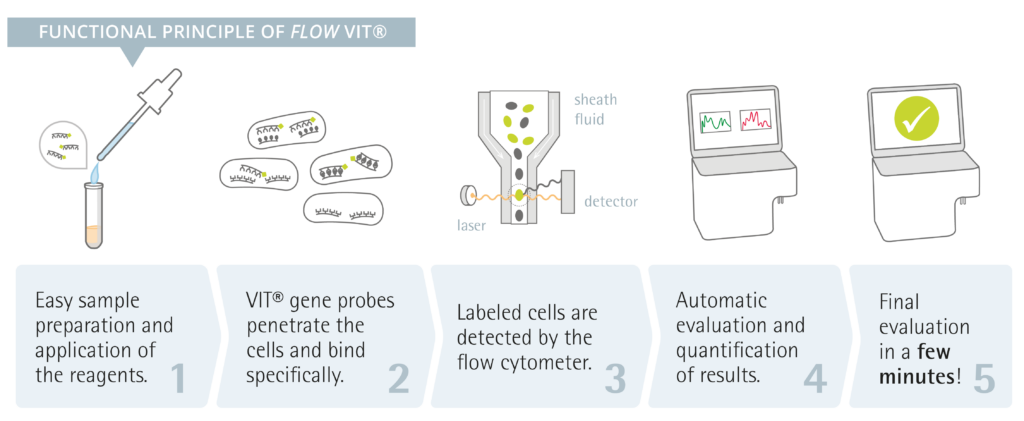
Flow-FISH is based on two significant pillars: Fluorescence In Situ Hybridization (FISH) and flow cytometry.
FISH is based on specific DNA oligonucleotides labeled with a fluorescence dye. Once they are introduced into bacterial cells by simple diffusion processes, they bind to their specific rRNA target sites within the bacterial species. The target sites within the ribosomes reflect the phylogeny of the bacteria and allow specific identification, as only the bacteria for which the oligonucleotides have been designed will attach to the specific probes. Once the oligonucleotides bind to their target sites, the dye will also be retained subsequently in the cell and cannot be washed away. As a result, the cells begin to shine when excited by a fluorescent light and can be identified. Moreover, it allows precise quantification and targets living cells, as only these have sufficient ribosomes to deliver a fluorescence signal.
Flow cytometry, on the other hand, is a powerful analytical technique used to measure certain properties of cells or particles as they pass through a laser beam in a fluid stream. The fundamental principle of flow cytometry involves hydrodynamically focusing the sample stream so that individual cells pass through a focused laser beam one at a time. As each cell intersects the laser, it scatters light and may emit fluorescence if it has been labeled with fluorescent markers. The scattered light and fluorescence signals are then collected by detectors, which convert them into electronic signals that are analyzed by a computer. Forward scatter (FSC) provides information about cell size, while side scatter (SSC) indicates cell granularity or internal complexity.
Flow-FISH combines FISH with flow cytometry so that only living cells can be specifically identified and reliably quantified. The examination can be automated and allows for an easy and practical workflow.
Comparison with Traditional Plate Count and Live/Dead Staining
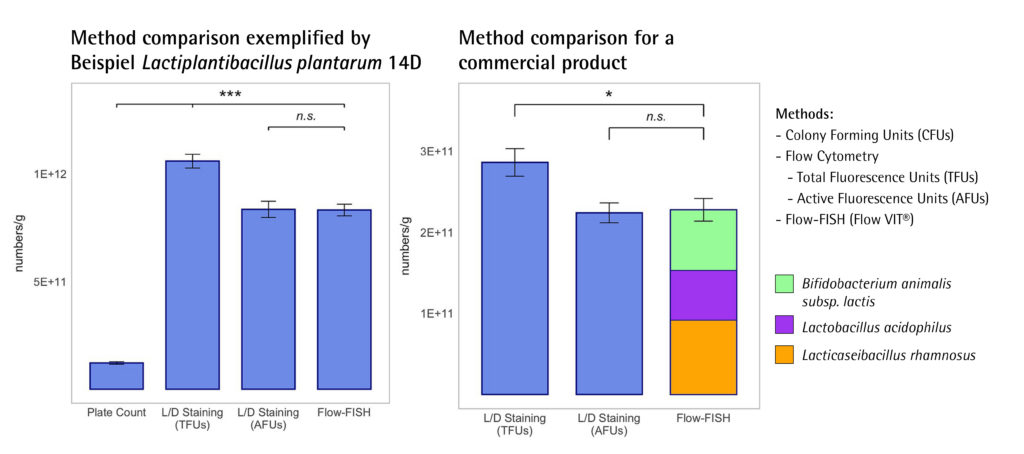
In the new study, the developed Flow-FISH method was compared with traditional plate count and Live/Dead staining methods for quantifying viable cells in probiotic products. The results demonstrated that Flow-FISH provided higher viable cell counts than plate count, challenging the traditional “gold standard” by revealing its tendency to underestimate actual viable cell numbers.
Compared to the results of Live/Dead staining, Flow-FISH delivered comparable results in accuracy and reliability, but with the huge difference that it could additionally specifically identify and quantify different bacterial species.
Application in Quality Control
The Flow-FISH technology can be applied to analyze probiotic products and is, therefore, an innovative tool for a new generation of reliable quality control for probiotics. By analyzing bacterial species in parallel, it delivers a detailed picture of which species and in what quantities they are viable in the product. The probiotic producer gets the following results:
- How many total bacteria are alive and how many are dead.
- The exact quantification of each viable bacterial species in their product.
This information is extremely valuable when assessing the efficiency of the production process and the quality of the probiotic product.
The Flow-FISH control can be applied as a service from vermicon as well as with commercial kits which target the most important probiotic bacteria. On this basis, over the next months, the portfolio of so-called Flow VIT® kits for probiotics bacteria will be continuously expanded.
Advantages of Flow-FISH in the Probiotic Industry
- Accurate Viable Cell Enumeration: Traditional plate count methods can significantly underestimate the number of viable cells due to the presence of VBNC cells. These metabolically active cells can contribute to the probiotic effect but do not form colonies on agar plates. Flow-FISH accurately counts these cells by targeting their rRNA, providing a true representation of viable cell counts.
- Specificity in Multi-Species Blends: Probiotic products often contain multiple bacterial species, each contributing uniquely to the overall health benefits. Flow-FISH enables the specific identification and quantification of each species within a blend, ensuring that the product meets its label claims.
- Enhanced Quality Control: With Flow-FISH, manufacturers can ensure consistent quality in their probiotic products. The method’s high sensitivity and specificity allow for precise monitoring of bacterial populations throughout the production process, from raw materials to the final product.
- Speed and Efficiency: The optimized Flow-FISH protocol significantly reduces analysis time and eliminates labor-intensive steps. This makes the method not only more accurate but also faster and easier to implement in a routine quality control setting.
Practical Demonstration in the Publication
In the published validation study, Flow-FISH was applied to analyze lyophilized samples of Lacticaseibacillus rhamnosus, Lactiplantibacillus plantarum, and Bifidobacterium animalis subsp. lactis, as well as a commercial probiotic product. The method demonstrated its capability to accurately enumerate viable cells, providing valuable data for quality control and product formulation.
Benefits of Flow-FISH for Different Stakeholders
Manufacturers benefit from the precision and speed of Flow-FISH. It allows for direct monitoring of probiotic cultures during production, ensuring that the final product meets the specified quality standards. This can lead to reduced wastage and more efficient production processes.
Regulatory bodies can use Flow-FISH as a reliable standard for assessing the viability and composition of probiotic products. The method’s accuracy and reproducibility make it an excellent tool for enforcing quality standards and protecting consumer health.
Researchers in the field of microbiology and probiotics can leverage Flow-FISH for more detailed studies on the viability and interactions of probiotic bacteria. The method’s ability to differentiate between species within a blend provides deeper insights into microbial dynamics and their health effects.
New Avenues with Flow-FISH
The application of Flow-FISH in the probiotic industry opens also new avenues for research and product development. It holds the potential to:
a) Improve Product Efficacy: By providing precise data on viable cell counts and species composition, Flow-FISH can help optimize probiotic formulations for better health outcomes.
b) Advance Microbiome Research: The method’s ability to accurately enumerate specific bacterial species makes it a valuable tool for studying the human gut microbiome and its interactions with probiotics.
c) Enhance Regulatory Standards: With its robust and reliable results, Flow-FISH can set new standards for probiotic quality control, ensuring that products are effective for consumers.
Conclusion
The development and implementation of Flow-FISH represents a major advancement in the field of microbiology and probiotics. This method not only improves the accuracy and reliability of viable cell enumeration but also enhances the overall quality control processes in the probiotic industry. It will undoubtedly lead to improvements in probiotic product quality and efficacy, ultimately benefiting both manufacturers and consumers alike.
Dr. Jiri Snaidr, vermicon AG, Zeppelinstr. 3, 85399 Hallbergmoos
The editorial staff of MicrobiomePost is not responsible for the text of this press release, which has been published in its entirety and without changes.