Introduction
The human skin is colonized by millions of microbes (i.e., bacteria, fungi, and viruses)1 that, together with their genetic information, constitutes the skin microbiome.
The study of the human skin microbiome – which has become a cornerstone in the cosmetic and dermatological fields2 – is broadening our understanding of skin health and disease and facilitating the development of topical pre/pro/postbiotics and live biotherapeutic products (LBPs) for different skin conditions.
Most skin microbiome studies currently use next-generation sequencing (NGS) approaches, specifically amplicon sequencing and shotgun metagenomics3. In this report, we will go through three key factors to consider when using skin microbiome profiling tools.
Resolution
Amplicon sequencing is the most common strategy to characterize cutaneous microbial communities. While this method has key strengths, such as its cost-effectiveness, it is usually limited to genus-level resolution4. A recent breakthrough in NGS platforms has been the development of long-read technologies, such as Oxford Nanopore. While full-length 16S rRNA gene amplicon sequencing has shown to give a better resolution at the species level than short-read technologies5, long-read platforms tend to have lower per base accuracy, higher total costs, and lower throughput6,7.
Shotgun metagenomics, which is increasingly used in skin microbiome profiling, allows for taxonomic identification at species and even strain level2. This high resolution is relevant when strains within a species harbor different gene content that determines functional differences1. For example, C. acnes strains have different pathogenic potential8. However, the “price” for the increased resolution of shotgun metagenomics is higher sequencing costs and more complex data analysis9. These become more relevant issues as the number of samples analyzed increases – for example, when profiling a high number of trial participants, monitoring probiotics/LBP engraftment on the skin, performing quality control of probiotics/LBP production runs, or assessing the effect of a skin product over time.
Host DNA and biomass
Amplicon sequencing, targeting the bacterial 16S rRNA gene and the fungal ITS1 region, can be used in samples containing host DNA10. As mentioned above, such an approach is generally only able to provide genus-level information.
Although shotgun metagenomics can provide important species and subspecies resolution, its use is challenging in skin samples. It requires a high starting amount of DNA11, which may pose a challenge for skin samples, which have a low microbial biomass12. In addition, skin samples contain high levels of host DNA, where it can represent more than 90% of the total extracted DNA3. Therefore, although host DNA sequences can be removed later during data processing, a large part of the reads have already been “spent” on sequencing the human genome. This results in increased costs due to the deep sequencing runs required to yield relevant information. The depletion of host DNA is another way to reduce human reads from shotgun metagenomics and consequently decrease sequencing costs. However, different host DNA-depletion approaches have shown taxonomic biases13.
Absolute Quantification
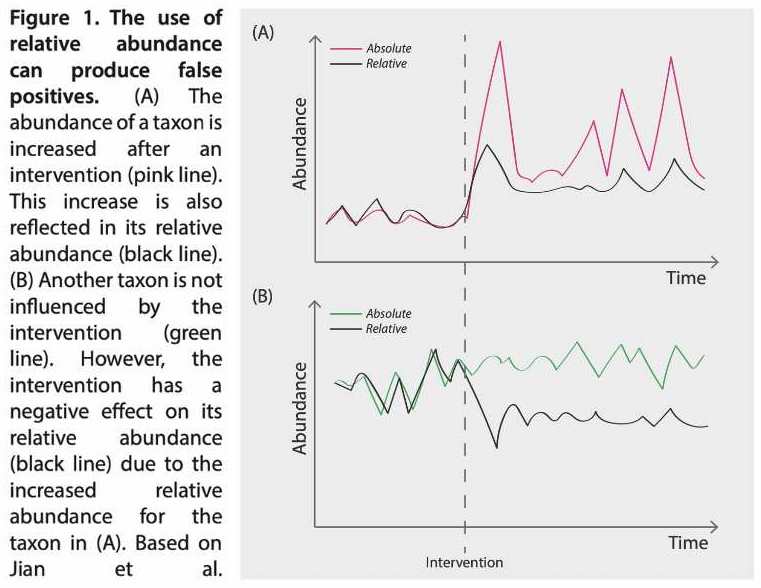
As NGS instruments have a fixed capacity (number of slots to fill), the data obtained for all detected taxa will be described as relative abundances – which are mutually dependent – but not as absolute numbers. This poses a challenge, as relative abundances can lead to misleading results14 (Figure 1). In addition, the quantification of skin microorganisms is important to address the bioburden of common skin microbes and whether it increases in specific skin disorders15.
Several approaches allow absolute quantification of microbial taxa, such as qPCR16. Different methods have been integrated into NGS pipelines to allow for quantitative data, e.g., exogenous spike-in bacteria8. However, they increase the cost and complexity of the data generated and are not yet widely used in the human microbiome field14.
Transforming Skin Microbiome Analysis with Precision Microbiome Profiling (PMP™)
Bio-Me has developed the Comprehensive Skin Microbiome panel with a precise analysis of 50 key bacterial and fungal targets. The PMP™ approach is built on a qPCR-based platform delivering accurate information about the microbiome in a high-throughput format. Results are directly actionable – no need for bioinformatics steps for data processing – and produced more rapidly (4 hours of turnaround time for 192 samples) and more cost-effectively than traditional sequencing approaches.
In addition, the Comprehensive Skin Microbiome panel meets the needs for a skin microbiome profiling tool:
- High-level resolution – It provides the same resolution as shotgun metagenomics (down to species and subspecies).
- Not impacted by host DNA – The targeted approach used overcomes the challenges associated with a high presence of host DNA in the sample.
- Quantitative – It provides the number of genomes per taxa, as well as the relative abundance for the targeted taxa.
Custom proprietary strains assay design upon request.
Of note, specific skin disorders have been linked to the status of the gut microbiome, and the gut-skin axis is an emerging research area17. Bio-Me has also developed a Gut Microbiome Extensive Panel consisting of close to 100 unique assays for specific gut bacterial and fungal targets.